surfinpy.mu_vs_mu¶
Functions related to the generation of surface phase diagrams as a function of chemical potential. An explanation of theory can be found here
- surfinpy.mu_vs_mu.calculate(data, bulk, deltaX, deltaY, x_energy=0, y_energy=0, increments=0.025)[source]¶
Initialise the surface energy calculation.
- Parameters
data (
list) – List ofsurfinpy.data.DataSetfor each phasebulk (
surfinpy.data.ReferenceDataSet) – Data for bulkdeltaX (
dict) – Range of chemical potential/label for species XDeltaY (
dict) – Range of chemical potential/label for species Yx_energy (
float) – DFT energy of adsorbing speciesy_energy (
float) – DFT energy of adsorbing species
- Returns
system – Plotting object
- Return type
- surfinpy.mu_vs_mu.calculate_excess(adsorbant, slab_cations, area, bulk, nspecies=1, check=False)[source]¶
Calculates the excess of a given species at the surface. Depending on the nature of the species, there are two ways to do this. If the species is a constituent part of the surface, e.g. Oxygen in
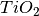 then the calculation must account for
the stoichiometry of that material. Using the
then the calculation must account for
the stoichiometry of that material. Using the 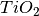 example
example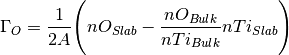
where
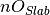 is the number of oxygen in the slab,
is the number of oxygen in the slab,
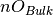 is the number of oxygen in the bulk,
A is the surface area,
is the number of oxygen in the bulk,
A is the surface area, 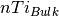 is the number of Ti in
the bulk and
is the number of Ti in
the bulk and 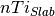 is the number of Ti in the slab.
If the species is just an external adsorbant, e.g. water or carbon dioxide
then one does not need to consider the state of the surface,
as there was none there to begin with.
is the number of Ti in the slab.
If the species is just an external adsorbant, e.g. water or carbon dioxide
then one does not need to consider the state of the surface,
as there was none there to begin with.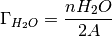
where
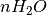 is the number of water molecules and A is the
surface area.
is the number of water molecules and A is the
surface area.- Parameters
adsorbant (
int) – Number of speciesslab_cations (
int) – Number of cationsarea (
float) – Area of surfacebulk (
dict) – Dictonary of bulk propertiesnspecies (
int) – number of external speciescheck (
bool) – Check if this is an external or constituent species.
- Returns
Surface excess of given species.
- Return type
float
- surfinpy.mu_vs_mu.calculate_normalisation(slab_energy, slab_cations, bulk, area)[source]¶
Normalises the slab energy relative to the bulk material. Thus allowing the different slab calculations to be compared.
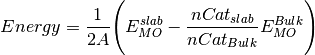
where Energy is the slab energy normalised to the bulk,
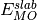 is the DFT slab energy,
is the DFT slab energy, 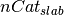
is the number of slab cations,
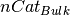 is the number of bulk
is the number of bulkcations,
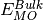 is the DFT bulk energy A is the surface
area.
is the DFT bulk energy A is the surface
area.- Parameters
slab_energy (
float) – Energy of the slab from DFTslab_cations (
int) – Total number of cations in the slabbulk (
surfinpy.data.DataSet) – Bulk propertiesarea (
float) – Surface area
- Returns
Constant normalising the slab energy to the bulk energy.
- Return type
float
- surfinpy.mu_vs_mu.calculate_surface_energy(deltamux, deltamuy, x_energy, y_energy, xexcess, yexcess, normalised_bulk)[source]¶
Calculates the surface for a given chemical potential of species x and species y for a single phase.
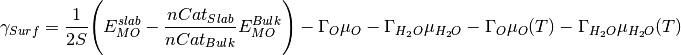
where S is the surface area,
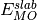 is the DFT energy of
the stoichiometric slab,
is the DFT energy of
the stoichiometric slab, 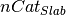 is the number of cations
in the slab,
is the number of cations
in the slab, 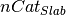 is the number of cations in the bulk
unit cell,
is the number of cations in the bulk
unit cell, 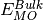 is the DFT energy of the bulk unit cell,
is the DFT energy of the bulk unit cell,
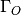
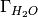 is the excess oxygen / water at
the surface and
is the excess oxygen / water at
the surface and 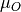
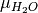 is the oxygen /
water chemcial potential.
is the oxygen /
water chemcial potential.- Parameters
deltamux (
array_like) – Chemical potential of species xdeltamuy (
array_like) – Chemical potential of species yx_energy (
float) – DFT energy or temperature corrected DFT energyy_energy (
float) – DFT energy or temperature corrected DFT energyxexcess (
float) – Surface excess of species xyexcess (
float) – Surface excess of species ynormalised_bulk (
float) – Slab energy normalised to the bulk value.
- Returns
2D array of surface energies as a function of chemical potential of x and y
- Return type
array_like
- surfinpy.mu_vs_mu.evaluate_phases(data, bulk, x, y, nsurfaces, x_energy, y_energy)[source]¶
Calculates the surface energies of each phase as a function of chemical potential of x and y. Then uses this data to evaluate which phase is most stable at that x/y chemical potential cross section.
- Parameters
data (
list) – List containing thesurfinpy.data.DataSetfor each phasebulk (
surfinpy.data.DataSet) – Data for bulkx (
dict) – X axis chemical potential valuesy (
dict) – Y axis chemical potential valuesnsurfaces (
int) – Number of phasesx_energy (
float) – DFT 0K energy for species xy_energy (
float) – DFT 0K energy for species y
- Returns
phase_data – array of ints, with each int corresponding to a phase.
- Return type
array_like